당뇨병 환자 예측 모델링
data shape : 2000000 x 28
# data analysis and wrangling
import pandas as pd
import numpy as np
import random as rnd
pd.set_option('display.max_columns', 100)
# visualization
import seaborn as sns
import matplotlib.pyplot as plt
%matplotlib inline
# machine learning
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC, LinearSVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import Perceptron
from sklearn.linear_model import SGDClassifier
from sklearn.tree import DecisionTreeClassifier
data_2017 = pd.read_csv('./NHIS_2017_2018_100m/NHIS_OPEN_GJ_2017_100.csv')
data_2018 = pd.read_csv('./NHIS_2017_2018_100m/NHIS_OPEN_GJ_2018_100.csv')
df = pd.concat([data_2017,data_2018])
drop_cols = ['HCHK_YEAR', 'IDV_ID', 'TTH_MSS_YN', 'ODT_TRB_YN', 'WSDM_DIS_YN', 'DATA_STD__DT']
df.drop(drop_cols, axis = 1, inplace = True)
df.head()
| SEX | AGE_GROUP | HEIGHT | WEIGHT | WAIST | SIGHT_LEFT | SIGHT_RIGHT | HEAR_LEFT | HEAR_RIGHT | BP_HIGH | BP_LWST | BLDS | TOT_CHOLE | TRIGLYCERIDE | HDL_CHOLE | LDL_CHOLE | HMG | OLIG_PROTE_CD | CREATININE | SGOT_AST | SGPT_ALT | GAMMA_GTP | SMK_STAT_TYPE_CD | DRK_YN | HCHK_OE_INSPEC_YN | CRS_YN | TTR_YN | SIDO_50 | SIDO_busan | SIDO_chongB | SIDO_chongN | SIDO_dagu | SIDO_dajeon | SIDO_gangwon | SIDO_gyeongB | SIDO_gyeongN | SIDO_gyeonggi | SIDO_incheon | SIDO_jeonB | SIDO_jeonN | SIDO_kwangju | SIDO_sejong | SIDO_seoul | SIDO_ulsan | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 8 | 170 | 75 | 90.0 | 1.0 | 1.0 | 1.0 | 1.0 | 120.0 | 80.0 | 0.0 | 193.0 | 92.0 | 48.0 | 126.0 | 17.1 | 1.0 | 1.0 | 21.0 | 35.0 | 40.0 | 1.0 | 1.0 | 1 | NaN | 1.0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 7 | 180 | 80 | 89.0 | 0.9 | 1.2 | 1.0 | 1.0 | 130.0 | 82.0 | 0.0 | 228.0 | 121.0 | 55.0 | 148.0 | 15.8 | 1.0 | 0.9 | 20.0 | 36.0 | 27.0 | 0.0 | 0.0 | 1 | NaN | 2.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 2 | 1 | 9 | 165 | 75 | 91.0 | 1.2 | 1.5 | 1.0 | 1.0 | 120.0 | 70.0 | 0.0 | 136.0 | 104.0 | 41.0 | 74.0 | 15.8 | 1.0 | 0.9 | 47.0 | 32.0 | 68.0 | 1.0 | 0.0 | 0 | NaN | NaN | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 1 | 11 | 175 | 80 | 91.0 | 1.5 | 1.2 | 1.0 | 1.0 | 145.0 | 87.0 | 0.0 | 201.0 | 106.0 | 76.0 | 104.0 | 17.6 | 1.0 | 1.1 | 29.0 | 34.0 | 18.0 | 1.0 | 0.0 | 1 | NaN | 0.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 1 | 11 | 165 | 60 | 80.0 | 1.0 | 1.2 | 1.0 | 1.0 | 138.0 | 82.0 | 0.0 | 199.0 | 104.0 | 61.0 | 117.0 | 13.8 | 1.0 | 0.8 | 19.0 | 12.0 | 25.0 | 1.0 | 0.0 | 1 | NaN | 0.0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Analyze by describing data¶
feature들의 타입을 확인한다.
- categorical
- numerical
print(df.columns.values)
['SEX' 'AGE_GROUP' 'SIDO' 'HEIGHT' 'WEIGHT' 'WAIST' 'SIGHT_LEFT'
'SIGHT_RIGHT' 'HEAR_LEFT' 'HEAR_RIGHT' 'BP_HIGH' 'BP_LWST' 'BLDS'
'TOT_CHOLE' 'TRIGLYCERIDE' 'HDL_CHOLE' 'LDL_CHOLE' 'HMG' 'OLIG_PROTE_CD'
'CREATININE' 'SGOT_AST' 'SGPT_ALT' 'GAMMA_GTP' 'SMK_STAT_TYPE_CD'
'DRK_YN' 'HCHK_OE_INSPEC_YN' 'CRS_YN' 'TTR_YN']
df.head()
| SEX | AGE_GROUP | SIDO | HEIGHT | WEIGHT | WAIST | SIGHT_LEFT | SIGHT_RIGHT | HEAR_LEFT | HEAR_RIGHT | BP_HIGH | BP_LWST | BLDS | TOT_CHOLE | TRIGLYCERIDE | HDL_CHOLE | LDL_CHOLE | HMG | OLIG_PROTE_CD | CREATININE | SGOT_AST | SGPT_ALT | GAMMA_GTP | SMK_STAT_TYPE_CD | DRK_YN | HCHK_OE_INSPEC_YN | CRS_YN | TTR_YN | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 8 | 43 | 170 | 75 | 90.0 | 1.0 | 1.0 | 1.0 | 1.0 | 120.0 | 80.0 | 99.0 | 193.0 | 92.0 | 48.0 | 126.0 | 17.1 | 1.0 | 1.0 | 21.0 | 35.0 | 40.0 | 1.0 | 1.0 | 1 | NaN | 1.0 |
| 1 | 1 | 7 | 11 | 180 | 80 | 89.0 | 0.9 | 1.2 | 1.0 | 1.0 | 130.0 | 82.0 | 106.0 | 228.0 | 121.0 | 55.0 | 148.0 | 15.8 | 1.0 | 0.9 | 20.0 | 36.0 | 27.0 | 3.0 | 0.0 | 1 | NaN | 2.0 |
| 2 | 1 | 9 | 41 | 165 | 75 | 91.0 | 1.2 | 1.5 | 1.0 | 1.0 | 120.0 | 70.0 | 98.0 | 136.0 | 104.0 | 41.0 | 74.0 | 15.8 | 1.0 | 0.9 | 47.0 | 32.0 | 68.0 | 1.0 | 0.0 | 0 | NaN | NaN |
| 3 | 1 | 11 | 48 | 175 | 80 | 91.0 | 1.5 | 1.2 | 1.0 | 1.0 | 145.0 | 87.0 | 95.0 | 201.0 | 106.0 | 76.0 | 104.0 | 17.6 | 1.0 | 1.1 | 29.0 | 34.0 | 18.0 | 1.0 | 0.0 | 1 | NaN | 0.0 |
| 4 | 1 | 11 | 30 | 165 | 60 | 80.0 | 1.0 | 1.2 | 1.0 | 1.0 | 138.0 | 82.0 | 101.0 | 199.0 | 104.0 | 61.0 | 117.0 | 13.8 | 1.0 | 0.8 | 19.0 | 12.0 | 25.0 | 1.0 | 0.0 | 1 | NaN | 0.0 |
categorical features
- SEX, SIDO
- HEAR_LEFT
- HEAR_RIGHT
- SMK_STAT_TYPE_CD
- DRK_YN
- HCHK_OE_INSPEC_YN
- CRS_YN, TTR_YN
numerical features
Continous
- HEIGHT
- WEIGHT
- WAIST, SIGHT_LEFT
- SIGHT_RIGHT
- BP_HIGH
- BP_LWST
- BLDS
- TOT_CHOLE
- TRIGLYCERIDE
- HDL_CHOLE
- LDL_CHOLE
- HMG
- CREATININE
- SGOT_AST
- SGPT_ALT
- GAMMA_GTP
Discrete
- AGE_GROUP
- OLIG_PROTE_CD
categorical = ['SEX', 'SIDO', 'HEAR_LEFT', 'HEAR_RIGHT', 'SMK_STAT_TYPE_CD', 'DRK_YN', 'HCHK_OE_INSPEC_YN', 'CRS_YN', 'TTR_YN']
numerical = ['HEIGHT', 'WEIGHT', 'WAIST', 'SIGHT_LEFT', 'SIGHT_RIGHT', 'BP_HIGH', 'BP_LWST', 'BLDS', 'TOT_CHOLE', 'TRIGLYCERIDE', 'HDL_CHOLE', 'LDL_CHOLE', 'HMG', 'CREATININE', 'SGOT_AST', 'SGPT_ALT', 'GAMMA_GTP', 'AGE_GROUP', 'OLIG_PROTE_CD']
null values
WAIST
SIGHT_LEFT
SIGHT_RIGHT
HEAR_LEFT
HEAR_RIGHT
BP_HIGH
BP_LWST
BLDS
TOT_CHOLE
TRIGLYCERIDE
HDL_CHOLE
LDL_CHOLE
HMG
OLIG_PROTE_CD
CREATININE
SGOT_AST
SGPT_ALT
GAMMA_GTP
SMK_STAT_TYPE_CD
DRK_YN
CRS_YN
TTR_YN
df.isnull().sum()
SEX 0
AGE_GROUP 0
SIDO 0
HEIGHT 0
WEIGHT 0
WAIST 680
SIGHT_LEFT 436
SIGHT_RIGHT 455
HEAR_LEFT 375
HEAR_RIGHT 373
BP_HIGH 5749
BP_LWST 5748
BLDS 5957
TOT_CHOLE 667287
TRIGLYCERIDE 667297
HDL_CHOLE 667298
LDL_CHOLE 677034
HMG 5966
OLIG_PROTE_CD 15162
CREATININE 5949
SGOT_AST 5947
SGPT_ALT 5948
GAMMA_GTP 5951
SMK_STAT_TYPE_CD 378
DRK_YN 355479
HCHK_OE_INSPEC_YN 0
CRS_YN 1600008
TTR_YN 1199484
dtype: int64
Distribution of datasets
df[numerical].describe()
| HEIGHT | WEIGHT | WAIST | SIGHT_LEFT | SIGHT_RIGHT | BP_HIGH | BP_LWST | BLDS | TOT_CHOLE | TRIGLYCERIDE | HDL_CHOLE | LDL_CHOLE | HMG | CREATININE | SGOT_AST | SGPT_ALT | GAMMA_GTP | AGE_GROUP | OLIG_PROTE_CD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 2.000000e+06 | 2.000000e+06 | 1.999320e+06 | 1.999564e+06 | 1.999545e+06 | 1.994251e+06 | 1.994252e+06 | 1.994043e+06 | 1.332713e+06 | 1.332703e+06 | 1.332702e+06 | 1.322966e+06 | 1.994034e+06 | 1.994051e+06 | 1.994053e+06 | 1.994052e+06 | 1.994049e+06 | 2.000000e+06 | 1.984838e+06 |
| mean | 1.622604e+02 | 6.342898e+01 | 8.134754e+01 | 9.786358e-01 | 9.762449e-01 | 1.225481e+02 | 7.606142e+01 | 1.006600e+02 | 1.956388e+02 | 1.335479e+02 | 5.680952e+01 | 1.131066e+02 | 1.425832e+01 | 8.633026e-01 | 2.614770e+01 | 2.601790e+01 | 3.739504e+01 | 1.056190e+01 | 1.095813e+00 |
| std | 9.310089e+00 | 1.262693e+01 | 1.106285e+01 | 6.073918e-01 | 6.047442e-01 | 1.459186e+01 | 9.954033e+00 | 2.418251e+01 | 3.918748e+01 | 1.038458e+02 | 1.929372e+01 | 3.656292e+01 | 1.583889e+00 | 4.986576e-01 | 2.501259e+01 | 2.678962e+01 | 5.061595e+01 | 2.860501e+00 | 4.390614e-01 |
| min | 1.250000e+02 | 2.500000e+01 | 1.000000e+00 | 1.000000e-01 | 1.000000e-01 | 5.800000e+01 | 2.700000e+01 | 2.300000e+01 | 6.000000e+00 | 1.000000e+00 | 1.000000e+00 | 1.000000e+00 | 5.000000e-01 | 1.000000e-01 | 1.000000e+00 | 1.000000e+00 | 1.000000e+00 | 5.000000e+00 | 1.000000e+00 |
| 25% | 1.550000e+02 | 5.500000e+01 | 7.450000e+01 | 7.000000e-01 | 7.000000e-01 | 1.120000e+02 | 7.000000e+01 | 8.900000e+01 | 1.690000e+02 | 7.400000e+01 | 4.600000e+01 | 8.900000e+01 | 1.320000e+01 | 7.000000e-01 | 1.900000e+01 | 1.500000e+01 | 1.600000e+01 | 8.000000e+00 | 1.000000e+00 |
| 50% | 1.600000e+02 | 6.000000e+01 | 8.100000e+01 | 1.000000e+00 | 1.000000e+00 | 1.210000e+02 | 7.600000e+01 | 9.600000e+01 | 1.930000e+02 | 1.080000e+02 | 5.500000e+01 | 1.110000e+02 | 1.430000e+01 | 8.000000e-01 | 2.300000e+01 | 2.000000e+01 | 2.300000e+01 | 1.100000e+01 | 1.000000e+00 |
| 75% | 1.700000e+02 | 7.000000e+01 | 8.800000e+01 | 1.200000e+00 | 1.200000e+00 | 1.310000e+02 | 8.200000e+01 | 1.050000e+02 | 2.200000e+02 | 1.600000e+02 | 6.500000e+01 | 1.350000e+02 | 1.540000e+01 | 1.000000e+00 | 2.900000e+01 | 3.000000e+01 | 4.000000e+01 | 1.300000e+01 | 1.000000e+00 |
| max | 1.900000e+02 | 1.400000e+02 | 9.990000e+02 | 9.900000e+00 | 9.900000e+00 | 2.730000e+02 | 1.850000e+02 | 8.720000e+02 | 2.386000e+03 | 9.490000e+03 | 8.110000e+03 | 5.119000e+03 | 2.500000e+01 | 9.800000e+01 | 9.999000e+03 | 7.210000e+03 | 9.990000e+02 | 1.800000e+01 | 6.000000e+00 |
df[categorical].describe(include='all')
| SEX | SIDO | HEAR_LEFT | HEAR_RIGHT | SMK_STAT_TYPE_CD | DRK_YN | HCHK_OE_INSPEC_YN | CRS_YN | TTR_YN | |
|---|---|---|---|---|---|---|---|---|---|
| count | 2.000000e+06 | 2.000000e+06 | 1.999625e+06 | 1.999627e+06 | 1.999622e+06 | 1.644521e+06 | 2.000000e+06 | 399992.000000 | 800516.000000 |
| mean | 1.466995e+00 | 3.369548e+01 | 1.032698e+00 | 1.031518e+00 | 1.607232e+00 | 6.956865e-01 | 4.002580e-01 | 0.215164 | 0.603340 |
| std | 4.989096e-01 | 1.267530e+01 | 1.778441e-01 | 1.747127e-01 | 8.176123e-01 | 4.601162e-01 | 4.899507e-01 | 0.410937 | 0.591625 |
| min | 1.000000e+00 | 1.100000e+01 | 1.000000e+00 | 1.000000e+00 | 1.000000e+00 | 0.000000e+00 | 0.000000e+00 | 0.000000 | 0.000000 |
| 25% | 1.000000e+00 | 2.700000e+01 | 1.000000e+00 | 1.000000e+00 | 1.000000e+00 | 0.000000e+00 | 0.000000e+00 | 0.000000 | 0.000000 |
| 50% | 1.000000e+00 | 4.100000e+01 | 1.000000e+00 | 1.000000e+00 | 1.000000e+00 | 1.000000e+00 | 0.000000e+00 | 0.000000 | 1.000000 |
| 75% | 2.000000e+00 | 4.300000e+01 | 1.000000e+00 | 1.000000e+00 | 2.000000e+00 | 1.000000e+00 | 1.000000e+00 | 0.000000 | 1.000000 |
| max | 2.000000e+00 | 5.000000e+01 | 2.000000e+00 | 2.000000e+00 | 3.000000e+00 | 1.000000e+00 | 1.000000e+00 | 1.000000 | 2.000000 |
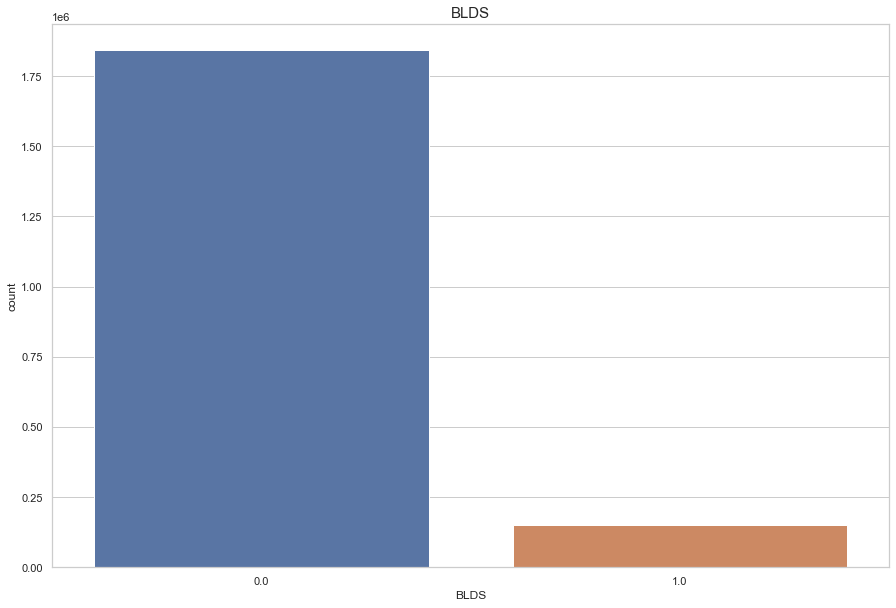
Target
BLDS
- 1 : 당뇨병
if BLDS >= 127 - 0 : 정상인
if BLDS < 127
Target은 결측치를 채워 넣을 수 없으니 결측치 drop 한다.
12.3 : 1 비율의 불균형 데이터
df.loc[ df['BLDS'] < 127, 'BLDS'] = 0
df.loc[ df['BLDS'] >= 127, 'BLDS'] = 1
df = df.dropna(subset = ['BLDS'], how = 'any', axis=0)
plt.figure(figsize=(15,10))
sns.countplot(df['BLDS'])
plt.title("BLDS",fontsize=15)
plt.show()
/Users/minsuha/anaconda3/lib/python3.8/site-packages/seaborn/_decorators.py:36: FutureWarning: Pass the following variable as a keyword arg: x. From version 0.12, the only valid positional argument will be `data`, and passing other arguments without an explicit keyword will result in an error or misinterpretation.
warnings.warn(